Harnessing the Power of Artificial Intelligence and Machine Learning to Boost CNS Drug Discovery

In recent years, there has been a surge of interest in the potential of artificial intelligence (AI) and machine learning (ML) tools, particularly deep learning models, to significantly reduce the time and cost associated with traditional drug discovery methods. By leveraging vast databases of chemical and biological information, these tools hold the promise of expediting the discovery of compounds with desired pharmacological profiles. To further enhance the speed and efficiency of existing drug discovery efforts for central nervous system disorders, we are exploring generative deep learning architectures for the de novo design of customized small molecules that employ transfer learning for pre-training and hybrid ligand-based and structure-based schemes for reinforcement learning. Additionally, we are developing transfer learning-enabled strategies for efficient in silico screening of ultra-large chemical libraries to identify ligands with desired pharmacological profiles using dense or graph convolutional neural network models.
Representative Publications
Provasi, D., Konovalov, K., Riina, N., Cullen, O., Filizola, M. “BOLD-GPCRs: A Transformer-Powered App for Predicting Ligand Bioactivity and Mutational Effects Across Class A GPCRs” (2025) bioRxiv doi: https://doi.org/10.1101/2025.08.04.668547
Provasi, D. and Filizola, M. “Fine-Tuned Deep Transfer Learning Models for Large Screenings of Safer Drugs Targeting Class A G Protein-Coupled Receptors” (2025) Biochemistry Mar 18;64(6):1328-1337. [PMID: 40056143]
Salas-Estrada L, Provasi D, Qui X, Kaniskan HÜ, Huang XP, DiBerto JF, Ribeiro JML, Jin J, Roth BL, Filizola M. De Novo Design of κ-Opioid Receptor Antagonists Using a Generative Deep Learning Framework; Journal of Chemical Information and Modeling (2023) Aug 28;63(16):5056-5065. [PMID: 37555591].
Integrative and Information-Driven Modeling of Biomolecular Complexes

The growing availability of cryo-EM structures of GPCRs and other membrane proteins, together with advances in proteomics, biophysics, and computation, has accelerated the adoption of integrative, information-driven modeling of biomolecular complexes. Our work employs a Bayesian integrative framework that couples computational data from metadynamics, the maximum-caliber principle, Markov state models, transition-path theory, and transfer-entropy analysis with experimental measurements from HDX-MS, TrIQ, smFRET, DEER, cryo-EM, and related techniques to place kinetics and allostery of GPCR-mediated transducer activation into structural context. We recently applied this framework to GPCR-mediated G-protein activation, integrating simulations with time-resolved experiments to uncover previously uncharacterized intermediates in β2-adrenergic-receptor signaling (in collaboration with K.Y. Chung, Sungkyunkwan University) and μ-opioid-receptor signaling (in collaboration with B.K. Kobilka, Stanford University, and G. Skiniotis, St. Jude Children’s Research Hospital). This broadly applicable approach continues to generate testable mechanistic hypotheses that inform rational drug design.
Representative Publications
Ahn, D., Provasi, D., Duc, N.M., Xu, J., Salas-Estrada, L., Spasic,, Yun, M.W., Kang, J., Gim, D., Lee, J., Du, Y., Filizola, M., Chung, K.Y. “Gαs slow conformational transition upon GTP binding and a novel Gαs regulator” (2023); iScience Apr 8; 26(5):106603. doi: 10.1016/j.isci.2023.106603. [PMID: 37128611 ]
Meral, D. , Provasi, D., Filizola, M. “An Efficient Strategy to Estimate Thermodynamics and Kinetics of G Protein-Coupled Receptor Activation Using Metadynamics and Maximum Caliber” bioRxiv; https://doi.org/10.1101/367888; (2018) Journal of Chemical Physics 149(22):224101 [PMID: 30553249].
Molecular Modeling and Enhanced Molecular Dynamics Simulations of GPCRs (Especially Opioid Receptors) and Other Membrane Proteins
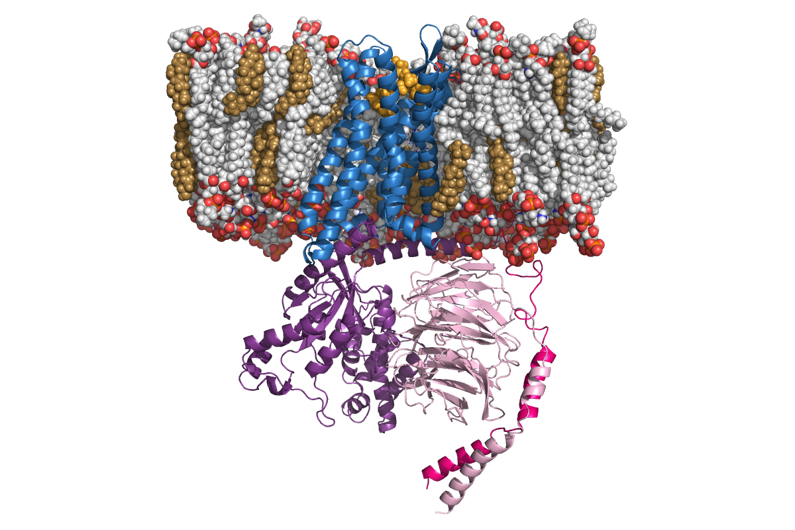
Our laboratory has advanced the use of enhanced molecular dynamics, coarse-grained and atomistic modeling, and machine learning to capture the conformational plasticity and long-timescale transitions of GPCRs and other membrane proteins that underlie drug action. These integrated approaches have enabled efficient mapping of ligand-recognition pathways and receptor-activation landscapes; rigorous estimation of thermodynamic and kinetic properties; and prediction of ligand-specific conformational states linked to functional selectivity.
A central focus of our work has been elucidating the molecular determinants of opioid receptor activation, biased signaling, and allosteric modulation. At a time when opioid pharmacology relied largely on static structural models, we introduced dynamic, atomistic descriptions of μ-opioid receptor conformational ensembles and activation pathways. Through enhanced sampling and ligand-specific simulations, we showed how agonists such as methadone and morphine stabilize distinct receptor states associated with divergent signaling outcomes, and we defined the molecular basis of positive allosteric modulation by small molecules and ions, helping shift the field toward a dynamics-centered view of GPCR pharmacology.
Representative Publications
Ribeiro Lamim, J.M., Provasi, D., Filizola, M. “A combination of machine learning and infrequent metadynamics to efficiently predict kinetic rates, transition states, and molecular determinants of drug dissociation from G protein-coupled receptors” Journal of Chemical Physics (2020) 153(12):124105
Kapoor, A., Provasi, D., Filizola, M. “Atomic-Level Characterization of the Methadone-Stabilized Active Conformation of µ-Opioid Receptor” (2020) Molecular Pharmacology, mol.119.119339.
Hu, X., Wang, Y., Hunkele, A., Provasi, D., Pasternak, G.W., Filizola, M. “Kinetic and thermodynamic insights into sodium ion translocation through the μ-opioid receptor from molecular dynamics and machine learning analysis” (2019) PLOS Computational Biology, 15(1):e1006689
Hu, X., Provasi, D., Filizola, M. “Mechanism of μ-Opioid Receptor-Magnesium Interaction and Positive Allosteric Modulation.” (2019) Biophysical Journal, pii: S0006-3495(19)30854-9
Provasi, D., Camacho-Artacho, M., Negri, A., Mobarec, J.C., Filizola, M. “Ligand-Induced Modulation of the Free-Energy Landscape of G Protein-Coupled Receptors Explored by Adaptive Biasing Techniques.” PLOS Computational Biology (2011) 7(10):e1002193.
Kapoor, A., Martinez-Rosell, Provasi, D., de Fabritiis, G., and Filizola, M. “Dynamic and Kinetic Elements of µ-Opioid Receptor Functional Selectivity” Scientific Reports, (2017) 7(1):11255.
Structure-Guided Drug Discovery and Chemotype Optimization
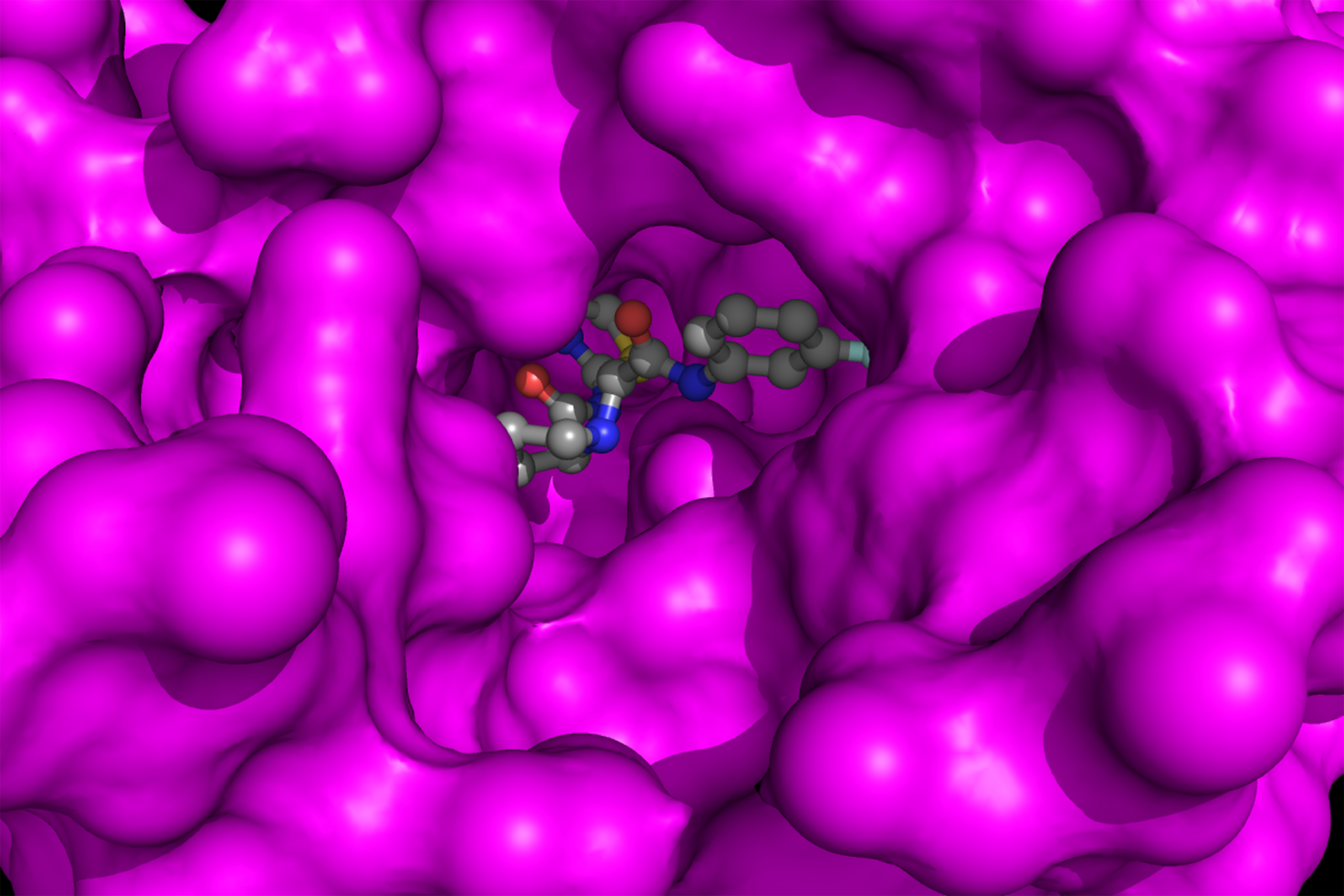
Our work has advanced structure- and computer-guided discovery of small-molecule modulators of GPCRs, integrins, ion channels, and oncology targets. Leveraging high-resolution structures, we integrated virtual screening, cheminformatics, molecular dynamics simulations, and generative AI with collaborative chemical synthesis, cryo-EM, and pharmacological studies. This multidisciplinary framework enabled the discovery of selective positive allosteric modulators of the δ- and μ-opioid receptors (δ/μOR) that enhance antinociceptive efficacy while minimizing adverse effects, mitragynine-derived μOR agonists with reduced liabilities, κOR receptor agonists and antagonists, a μOR–δOR heteromer-biased agonist with antinociceptive activity, small-molecule inhibitors for mantle cell lymphoma, αIIbβ3 and αVβ3 integrin antagonists that prevent receptor priming, TRPV5 channel inhibitors, and other first-in-class chemical series. These efforts demonstrated that computation-informed design can prospectively shape signaling bias, safety profiles, and mechanismspecific pharmacology, providing leads for therapeutic development in pain, thrombosis, cancer, and channelopathies. This work has been advanced through close collaborations with numerous colleagues, including: A. Alt & J.Traynor, U. Michigan; M. Canals, U. Nottingam; A.Christopoulos, Monash U.; B. Roth, UNC; Michael Cameron, Scripps Florida; Venetia Zachariou, Boston U.; D.Sames & J.A. Javitch, Columbia U.; S.Majumdar & T. Che, Washington U.; T. Prisinzano, Kansas U.; L. Devi, A. Aggarwal, J. Jin & S. Parekh, ISMMS; B.S. Coller, Rockefeller U.; V. Moinseenkova-Bell, U. Penn; T. Rohacs, Rutgers U.
Representative Publications
Sen, S., Spasic, A., Sinha, A., Wang, J., Bush, M., Li, J., Nešić, D., Zhou, Y., Angiulli, G., Morgan, P., Salas-Estrada, L., Takagi, J., Walz, T., Coller, B., Filizola, M. “Structure-Based Discovery of a Novel Class of Small-Molecule Pure Antagonist of Integrin aVb3” (2022) Journal of Chemical Information and Modeling Oct 24. doi: 10.1021/acs.jcim.2c00999.
Chakraborty, S., Diberto, J., Faouzi, A., Bernhard, S., Gutridge, A., Ramsey, S., Zhou, Y., Provasi, D., Nuthikattu, N., Jilakia, R., Nelson, M.N.F., Asher, W.B., Eans, S. O., Wilson, L.L., Chintala, S.M., Filizola, M., van Rijn, R.M., Margolis, E.B., Roth, B.L., McLaughlin, J.P., Che, T., Sames, D., Javitch, J.A., Majumdar, S. “A novel mitragynine analog with low efficacy mu-opioid receptor agonism displays antinociception with attenuated adverse effects” (2021) Journal of Medicinal Chemistry Sep 23;64(18):13873-13892.
Jatiani, S.S., Christie, S., Leshchenko, V., Jain, R., Kapoor, A., Bisignano, P., Lee, C., Kaniskan, H.U., Edwards, D., Meng, F., Lagana, A., Youssef, Y., Wiestner, A., Alinari, L., Jin, J., Filizola, M., Aggarwal, A. K., Parekh, S. “SOX11 Inhibitors Are Cytotoxic in Mantle Cell Lymphoma.” Clinical Cancer Research (2021) Aug 15;27(16):4652-4663.
Hughes, T.E.T., Del Rosario, J.S., Kapoor, A., Yazici, A. T., Fluck, E.C., Filizola, M., Rohacs, T., Moiseenkova-Bell, V.Y. “Structure-based discovery of novel TRPV5 inhibitors” (2019) eLife; pii: e49572. doi: 10.7554/eLife.49572.
Li, J., Fukase, Y., Shang, Y., Zou, W., Munoz-Felix, J., Buitrago, L., van Agthoven, J., Zhang, X., Hara, R., Tanaka,Y., Okamoto, R., Yasui, T., Nakahata, T., Imaeda, T., Aso, K., Zhou, Y., Locuson, C., Nesic, D., Duggan, M., Takagi, J., Vaughan, R., Walz, T., Hodivala-Dilke, K., Teitelbaum, S.L., Arnaout, A.M., Filizola, M., Foley, M.A., Coller, B.S. “Novel Pure αVβ3 Integrin Antagonists That Do Not Induce Receptor Extension, Prime the Receptor, or Enhance Angiogenesis at Low Concentrations” ACS Pharmacological & Translational Science (2019) 2, 6, 387-401.
Crowley, R.S., Riley, A.P., Sherwood, A.M., Groer, C.E., Shivaperumal, N., Biscaia, M., Paton, K., Schneider, S., Provasi, D., Kivell, B.M., Filizola, M., and Prisinzano, T.E. “Synthetic Studies of Neoclerodane Diterpenes from Salvia divinorum: Identification of a Potent and Centrally Acting μ Opioid Analgesic with Reduced Abuse Liability” (2016) Journal of Medicinal Chemistry, 59(24):11027-11038
Wardman, J.H., Gomes, I., Bobeck, E.N., Stockert, J., Kapoor, A., Bisignano, P., Gupta, A., Mezei, M., Kumar, S., Filizola, M., Devi, L.A. “Identification of a small molecule ligand that activates the neuropeptide receptor GPR171 and increases food intake” (2016) Science Signaling 9(430):ra55.
Kruegel, A.C., Gassaway, M.M., Kapoor, A., Varadi, A., Majumdar, S., Filizola, M., Javitch, J.A., Sames, D. “Synthetic and Receptor Signaling Explorations of the Mitragyna Alkaloids: Mitragynine as an Atypical Molecular Framework for Opioid Receptor Modulators” (2016) Journal of American Chemical Society 138(21):6754-64
Bisignano, P., Burford, N.T., Shang, Y., Marlow, B., Livingston, K.E., Fenton, A.M., Rockwell,K., Budenholzer, L., Traynor, J., Gerritz, S.W., Alt, A., and Filizola, M. “Ligand-Based Discovery of a New Scaffold for Allosteric Modulation of the mu-Opioid Receptor” (2015) Journal of Chemical Information & Modeling 55(9):1836-43
Li, J., Vootukuri, S., Shang, Y., Negri, A., Jiang, J.-K., Nedelman, M., Diacovo, T.G., Filizola, M., Thomas, C.J., Coller, B.S. “RUC-4: A Novel αIIbβ3 Antagonist for Pre-hospital Therapy of Myocardial Infarction” Arteriosclerosis, Thrombosis, and Vascular Biology (2014) Aug 21. pii: ATVBAHA.114.303724.
Gomes, I., Fujita, W., Gupta, A., Saldanha, A.S., Negri, A., Pinello, C.E., Roberts, E., Filizola, M., Hodder, P., and Devi, L.A. “Identification of a μOR-δOR heteromer-biased agonist with antinociceptive activity” Proc. Natl. Acad. Sci. USA (2013) 110(29):12072-7.
Zhu, J., Choi, W.-S., McCoy, J.G., Negri, A., Zhu, J., Naini, S., Li, J., Shen, M., Huang, W., Bougie, D., Rasmussen, M., Aster, R., Thomas, C.J., Filizola, M., Springer, T.A., and Coller, B.S. “Structure-Guided Design of a High Affinity Platelet Integrin αIIbβ3 Receptor Antagonist That Disrupts Mg2+ Binding to the MIDAS” Science Translational Medicine (2012) 4(125):1-13.
Burford, N., Livingston, K., Canals, M., Ryan, M., Budenholzer, L., Han, Y., Shang, Y., Herbst, J.J., O’Connell, J., Banks, M., Zhang, L., Filizola, M., Bassoni, D., Wehrman, T., Christopoulos, A., Traynor, J., Gerritz, S., Alt, A. “Discovery, Synthesis and Molecular Pharmacology of Selective Positive Allosteric Modulators of the δ-Opioid Receptor” (2015) Journal of Medicinal Chemistry Apr 22.
Negri, A., Rives, M.L., Caspers, M.J., Prisinzano, T.E., Javitch, J.A., and Filizola, M. “Discovery of a Novel Selective Kappa-Opioid Receptor Agonist Using Crystal Structure-Based Virtual Screening” Journal of Chemical Information and Modeling (2013) 53: 521-526.
Mechanistic Insights into GPCR Oligomerization

As evidence emerged that GPCRs may function as oligomeric assemblies, the structural and mechanistic basis of receptor–receptor interactions remained unclear. Our laboratory established rigorous multiscale computational models to define dimerization interfaces, relative stability, kinetics, and membrane-dependent spatial organization of GPCR assemblies. These studies differentiated transient from stable oligomers, demonstrated how transmembrane interactions tune receptor activation, and provided mechanistic frameworks to interpret receptor cross-talk and cooperative signaling. This work helped shift the conceptualization of GPCR signaling from isolated monomers to dynamic higher-order organizations and informed experimental design in living-cell imaging and pharmacological studies. I led the modeling and interpretation of these systems, integrating simulations with collaborative experimental validation (with J. Levitz, Weill Cornell Medicine; J.A. Javitch, Columbia U.; M.J. Lohse, Würzburg U.) to advance understanding of oligomer-dependent receptor function.
Representative Publications
Thibado, J.K., Tano, J.Y., Lee, J. Salas-Estrada, L., Provasi, D., Strauss, A., Ribeiro, J.M.L., Xiang, G., Broichhagen, J., Filizola, M., Lohse, M.L., Levitz, J., “Differences in interactions between transmembrane domains tune the activation of metabotropic glutamate receptors” eLife (2021) Apr 21;10:e67027.
Walsh, S., Mathiasen, S., Christensen, S.M., Fay, J.F., King, C., Provasi, D., Borrero, E., Rasmussen, S.G.F., Fung, J.J., Filizola, M., Hristova, K., Kobilka, B., Farrens, D.L., Stamou, D. “Single proteoliposome high content analysis reveals differences in the homo-oligomerization of GPCRs” (2018) Biophysical Journal 115(2):300-312.
Meral, D., Provasi, D., Prada-Gracia, D., Möller, J., Marino, K., Lohse, M.J., and Filizola, M. “Molecular details of dimerization kinetics reveal negligible populations of transient µ-opioid receptor homodimers at physiological concentrations” (2018) Scientific Reports 8(1):7705
Marino, K., Prada-Gracia, D., Provasi, D., Filizola, M. “Impact of Lipid Composition and Receptor Conformation on the Spatio-Temporal Organization of mu-Opioid Receptors in a Multi-component Plasma Membrane Model” (2016) PLOS Computational Biology 12(12):e1005240
Provasi, D., Boz, M.B., Johnston, J.M., Filizola, M. “Preferred Supramolecular Organization and Dimer Interfaces of Opioid Receptors from Simulated Self-Association” (2015) PLOS Computational Biology Mar 30;11(3):e1004148.
Johnston, J.M., Wang, H., Provasi, D., Filizola, M. “Assessing the Relative Stability of Dimer Interfaces in G Protein-Coupled Receptors.” PLOS Computational Biology (2012) 8(8): e1002649
Structure-Guided Mechanistic Advances in Integrin Biology and Antagonist Design
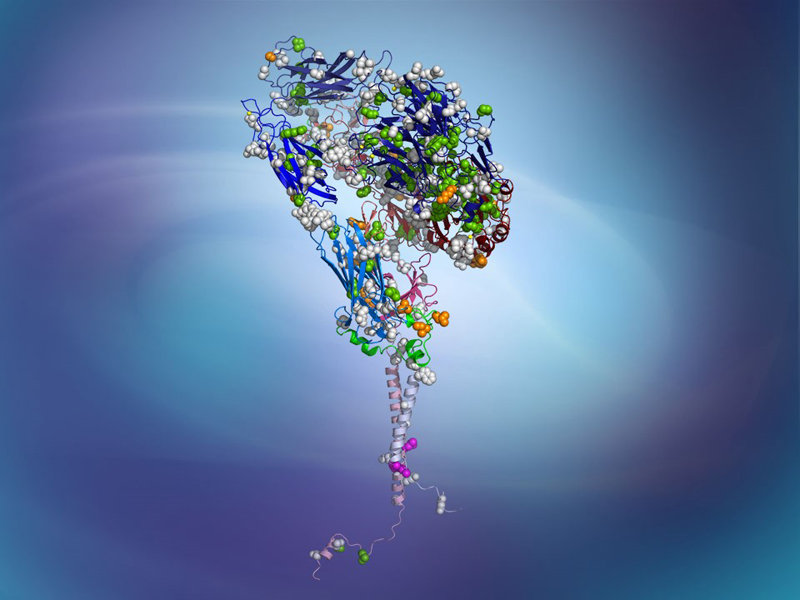
Through a long-standing collaboration with B.S. Coller at Rockefeller U., our laboratory contributed to elucidating the structural mechanisms linking ligand binding, metal coordination, conformational rearrangements, and activation of αIIbβ3 and αVβ3 integrins. Using structure-based modeling and molecular simulations, we defined molecular determinants that restrict receptor extension, clarified conformational changes associated with hybrid-domain swing-out, identified ancillary ligand-binding sites, and guided the discovery of pure antagonists that stabilize integrins in their resting state without priming or enhancing angiogenesis. These studies provided mechanistic foundations for the development of safer integrin-targeted therapeutics and informed interpretation of disease-associated variants emerging from next-generation sequencing. Notably, this work contributed to the discovery of the novel antiplatelet agent RUC-4 (zalunfiban), now in Phase III clinical trials as a single subcutaneous treatment for pre-hospital management of ST-Elevation Myocardial Infarction (STEMI).
Representative Publications
Sen, S., Spasic, A., Sinha, A., Wang, J., Bush, M., Li, J., Nešić, D., Zhou, Y., Angiulli, G., Morgan, P., Salas-Estrada, L., Takagi, J., Walz, T., Coller, B., Filizola, M. “Structure-Based Discovery of a Novel Class of Small-Molecule Pure Antagonist of Integrin aVb3” (2022) Journal of Chemical Information and Modeling Oct 24. doi: 10.1021/acs.jcim.2c00999.
Nešić, D., Bush, M., Spasic, A., Li, J., Kamata, T., Handa, M., Filizola, M., Thomas Walz, T., Coller, B.S., “Electron Microscopy of the αIIbβ3–PT25-2 Fab Complex Identifies a Mechanism by Which PT25-2 Induces αIIbβ3 Ligand Binding” (2021) Blood Advances, 5(7):1781-1790. [PMID: 33760023]
Nesic, D., Zhang, Y., Spasic, A., Li, J., Provasi, D., Filizola, M., Walz, T., and Coller, B.S. “Cryo-Electron Microscopy Structure of the αIIbβ3-Abciximab Complex” (2020) Arteriosclerosis, Thrombosis, and Vascular Biology 40(3):624-637.
Li, J., Fukase, Y., Shang, Y., Zou, W., Munoz-Felix, J., Buitrago, L., van Agthoven, J., Zhang, X., Hara, R., Tanaka,Y., Okamoto, R., Yasui, T., Nakahata, T., Imaeda, T., Aso, K., Zhou, Y., Locuson, C., Nesic, D., Duggan, M., Takagi, J., Vaughan, R., Walz, T., Hodivala-Dilke, K., Teitelbaum, S.L., Arnaout, A.M., Filizola, M., Foley, M.A., Coller, B.S. “Novel Pure αVβ3 Integrin Antagonists That Do Not Induce Receptor Extension, Prime the Receptor, or Enhance Angiogenesis at Low Concentrations” ACS Pharmacology & Translational Science (2019) 2, 6, 387-401.
Zafar, H., Shang, Y., Li, J., David, G.A. III, Fernandez, J.P., Molina, H., Filizola, M., Coller, B.S. “αIIbβ3-Binding to a Fibrinogen Fragment Lacking the g-chain Dodecapeptide is Activation Dependent and EDTA-Inducible (2017) Blood Advances 1:417-428.
Buitrago, L., Rendon, A., Liang, Y., Turro, E., Simeoni, I., Negri, A., ThromboGenomics Consortium, Filizola, M., Ouwehand, W.H., Coller, B.S. “αIIbβ3 Variants Defined by Next Generation Sequencing: Predicting Variants Likely to Cause Glanzmann Thrombasthenia” (2015) Proceedings of the National Academy of Science USA, 112(15):E1898-907.
Zhu, J., Choi, W.-S., McCoy, J. G., Negri, A., Zhu, J., Naini, S., Li, J., Shen, M., Huang, W., Bougie, D., Rasmussen, M., Aster, R., Thomas, C. J., Filizola, M., Springer, T. A., Coller, B. S. “Structure-Guided Design of a High-Affinity Platelet Integrin αIIbβ3Receptor Antagonist That Disrupts Mg2+ Binding to the MIDAS.” Science Translational Medicine (2012) 4: 125ra32
Negri, A., Li, J., Naini, S., Coller, B.S., Filizola, M. “Structure-Based Virtual Screening of Small-Molecule Antagonists of Platelet Integrin αIIbβ3that Do Not Prime the Receptor to Bind Ligand” Journal of Computer-Aided Molecular Design (2012) 26 (9): 1005-1015
Zhu, J., Zhu, J., Negri, A., Provasi, D., Filizola, M., Coller, B.S., Springer, T.A. “Closed headpiece of integrin αIIbβ3 and its complex with an αIIbβ3-specific antagonist that does not induce opening” Blood (2010) 116 (23):5050-5059.
Contributions to Team-Science Projects

By integrating state-of-the-art theoretical methods and advanced molecular simulations, our lab provides high-resolution mechanistic insight into biological systems that is often inaccessible through experiment alone. These computational discoveries both complement and guide experimental investigations, forming the basis for new hypotheses, targeted mechanistic studies, and innovative therapeutic strategies. Our work is deeply collaborative and has been strengthened through productive partnerships with leading investigators, including Dan Minor, UCSF; Sudha Chakrapani, Case Western Reserve University, and Jonathan A. Javitch, Columbia University, among others, enabling truly integrative, multidisciplinary progress at the interface of computation and experiment.
Representative Publications
Zakrzewska, S., Nixon, S.A., Chen, Z., Hajare, H.S., Park, E.R., Mulcahy, J.V., Arlinghaus, K.M., Neu, E., Konovalov, K., Provasi, D., Leighfield, T.A., Filizola, M., Du Bois, J., Minor, D. L. Jr. “Structural basis for saxitoxin congener binding and neutralization by anuran saxiphilins” BIORXIV/2024/616181 (2025) Nature Communications Apr 24;16(1):3885. [PMID: 40274765]
Basak, , Kumar, A. Ramsey, S., Gibbs, E., Kapoor, A., Filizola, M., Chakrapani, S. “High-resolution structures of multiple 5-HT3AR-setron complexes reveal a novel mechanism of competitive inhibition” elife (2020) 9:e57870.
Basak, S., Gicheru, Y., Kapoor, A., Mayer, M.L., Filizola, M. and Chakrapani, S. “Molecular mechanism of setron-mediated inhibition of full-length 5-HT3A receptor” (2019) Nature Communications 10(1): 3225
Yano, H., Provasi, D., Sheng Cai, N., Filizola, M., Ferré, S., Javitch, J.A., “Development of novel biosensors to study receptor-mediated activation of Gs and Golf” (2017) Journal of Biological Chemistry Dec 8;292(49):19989-19998.
Coudray, N., Valvo, S., Hu, M., Lasala, R., Kim, C., Vink, M., Zhou, M., Provasi, D., Filizola, M., Tao, J., Fang, J., Penczek, P.A., Ubarretxena-Belandia, I., Stokes, D.L. “Inward-Facing Conformation of the Zinc Transporter YiiP revealed by Cryo-electron Microscopy” Proc. Natl. Acad. Sci. USA (2013) 110(6):2140-2145.